Mastering AQA A Level Biology Section 3.4.2: DNA and Protein Synthesis - Common Questions & Mark Scheme Insights
After analyzing past papers and mark schemes for AQA specification section 3.4.2 (DNA and Protein Synthesis), I've identified the question types that consistently challenge students. Understanding these patterns and the specific language that mark schemes reward is essential for maximizing your exam performance. Let me guide you through four of the most frequently tested question types with real AQA examples.
Question Type 1: Comparing DNA and RNA Structures
Why this question type is common: This tests your ability to make precise comparisons between two fundamental molecules. Examiners use this format to assess whether you can distinguish structural features clearly and express them comparatively.
How to approach it (4 marks available):
The mark scheme requires direct comparisons - you must write features opposite each other:
"DNA has deoxyribose, mRNA has ribose"
"DNA has thymine, mRNA has uracil"
"DNA long, mRNA short"
"DNA is double stranded, mRNA is single stranded"
Alternative acceptable comparisons:
"DNA has hydrogen bonds, mRNA has no hydrogen bonds"
"DNA has (complementary) base pairing, mRNA does not"
Mark scheme insight: The mark scheme is very specific - "Must be comparisons." If you only write half of each comparison, you won't earn the mark. Write "DNA double helix" for 'double stranded' and "mRNA single helix" for 'single stranded' - both are acceptable. The mark scheme also notes to ignore references to splicing/introns for this particular question.
Common mistake: Students often write features in isolation rather than as comparisons. "DNA has deoxyribose" alone won't earn a mark - you need "DNA has deoxyribose, mRNA has ribose."
Question Type 2: Transcription Process Description
Why this question type is common: Transcription is a fundamental process that appears repeatedly. This question tests your ability to describe a sequence of events accurately while following specific constraints.
Step-by-step approach (3 marks available):
"(Free RNA) nucleotides form complementary base pairs" (1 mark)
"Phosphodiester bonds form" (1 mark)
"By (action of) RNA polymerase" (1 mark)
Mark scheme insight: Notice the specific exclusions - "Do not include DNA helicase or splicing." Follow these instructions precisely. The mark scheme accepts "A-U, G-C OR combination of those pairs" for complementary base pairing, and you can write "linkages" instead of "bonds" for phosphodiester bonds. However, you must mention RNA polymerase to earn the third mark.
Common mistake: Students often write about DNA helicase breaking hydrogen bonds or mention splicing, losing marks for not following the question constraints. Always read what you're told NOT to include.
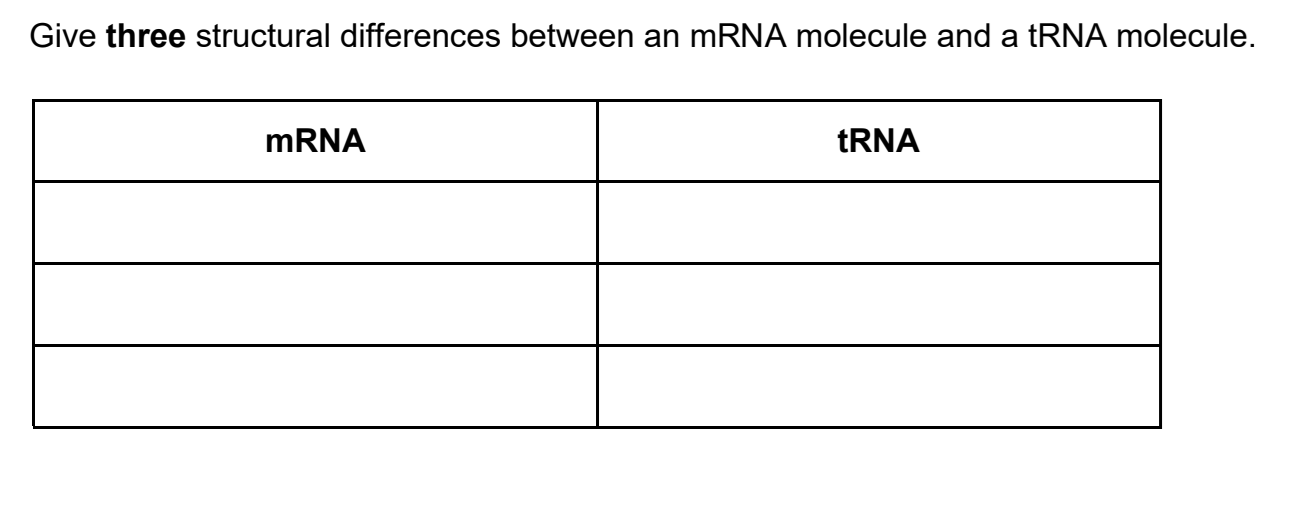
Question Type 3: Translation and tRNA Structure
Why this question type is common: This tests detailed knowledge of molecular structures involved in protein synthesis. It's perfect for discriminating between students who have memorized features and those who understand comparative structure.
How to structure your answer (3 marks available):
The mark scheme requires comparisons between mRNA and tRNA:
"mRNA (Has) codon(s) / tRNA (Has) anticodon"
"mRNA No hydrogen/H bonds/base pairs / tRNA Has hydrogen/H bonds/base pairs"
"mRNA No amino acid binding site / tRNA Has amino acid binding site"
"mRNA Linear/straight/not folded / tRNA 'Clover (leaf' shape)/folded"
"mRNA Long/many nucleotides/bases / tRNA Short/few nucleotides/bases"
Choose any three comparisons from this list.
Mark scheme insight: The mark scheme explicitly states "Must be comparisons" and accepts descriptions of binding sites (e.g., "amino acid only bound to tRNA" or "mRNA cannot carry an amino acid, tRNA can"). You can also write "CCA end" for amino acid binding site. Notice how precise the acceptable alternatives are - the mark scheme rewards accurate biological terminology.
Common mistake: Writing "tRNA is double stranded" is specifically rejected by the mark scheme. While tRNA has base pairing in its clover leaf structure, it's not considered double stranded like DNA.
Question Type 4: Gene Mutations and Functional Effects
Why this question type is common: This question type assesses understanding at multiple levels - from molecular changes to functional consequences. It's excellent for testing whether students can link DNA changes to protein function through multiple pathways.
How to structure your answer (4 marks available):
Possible explanations include:
"Substitution (mutation occurred)" (1 mark)
"(Only) one nucleotide/base pair is changed (in a gene)" OR "(Only) one (DNA) triplet/codon changed" (1 mark)
"Same amino acid (coded for)" (1 mark)
"(Because) DNA/genetic code is degenerate" (1 mark)
"(So) tertiary structure is not changed" (1 mark)
"(Change) could be in an intron" (1 mark)
"Removed during splicing" (1 mark)
Mark scheme insight: Maximum 4 marks, so you need to select the most relevant points. The mark scheme accepts descriptions of degenerate code and notes that marks 3 and 4 "can be awarded together, e.g 'different codons/triplets code for the same amino acid' = MP3 and MP4." This means a well-phrased sentence can earn multiple marks. The mark scheme rejects "same amino acid is produced" but accepts "same amino acid coded for" - subtle but important distinction. It also accepts "one amino acid changed" for mark point 3.
Multiple pathways to a correct answer:
Degenerate code pathway: substitution → same amino acid → no change in tertiary structure
Intron pathway: mutation in intron → removed during splicing → functional protein unchanged
Minor change pathway: one amino acid changed → tertiary structure unaffected
Common mistake: Students often describe the mutation but fail to explain why the protein remains functional. Link the molecular change to the functional consequence.
General Tips for Section 3.4.2 Success
Master comparative language: Questions often require direct comparisons. Practice writing features in parallel for DNA/RNA, mRNA/tRNA, prokaryotes/eukaryotes.
Follow exclusions religiously: When questions say "Do not include..." they mean it. Mark schemes penalize students who ignore these constraints.
Link molecular to functional: Don't just describe what happens - explain why it matters. Connect DNA changes → amino acid changes → protein structure → protein function.
Learn mark scheme synonyms: The mark scheme lists acceptable alternatives. For example:
"Bonds" = "linkages"
"Complementary base pairing" = "hydrogen bonding between bases"
"Folded" = "clover leaf shape" (for tRNA)
Use precise terminology: The mark scheme distinguishes between similar phrases:
"Same amino acid coded for" ✓
"Same amino acid is produced" ✗
Practice process descriptions: For transcription and translation, learn the sequence of events and the enzymes/molecules involved. Mark schemes reward step-by-step accuracy.
Understand degenerate code implications: Many mutation questions hinge on understanding that multiple codons code for the same amino acid. This explains why many mutations don't change protein function.
Key Concepts to Master
Transcription differences: Eukaryotes produce pre-mRNA that requires splicing; prokaryotes don't. This appears repeatedly in questions comparing the two systems.
Translation mechanics: Know the roles of:
mRNA (carries genetic code)
tRNA (brings specific amino acids, has anticodons)
Ribosomes (site of translation)
ATP (provides energy for peptide bond formation and amino acid-tRNA binding)
Structural comparisons: Be able to compare:
DNA vs RNA (sugar, bases, strands, length)
mRNA vs tRNA (shape, function, base pairing, length)
Prokaryotic vs eukaryotic protein synthesis (location, splicing, complexity)
Mutation effects: Understand why mutations may have:
No effect (degenerate code, introns, conservative substitutions)
Negative effects (frameshift, active site changes, nonsense mutations)
Positive effects (improved protein function, evolutionary advantages)
Remember, the mark scheme is your friend. It shows exactly what examiners want to see. Practice using mark scheme language in your answers, and you'll find your marks improving significantly. The key is precision - vague biological statements rarely earn marks, while specific, accurate terminology consistently does.
Section 3.4.2 builds on section 3.4.1's foundation, so ensure you're solid on DNA structure before tackling protein synthesis mechanisms. When you understand both the molecular details and the bigger picture of how genetic information flows from DNA → RNA → protein, even complex questions become manageable.
Good luck with your revision!